Update README.md
Browse files
README.md
CHANGED
|
@@ -1,3 +1,112 @@
|
|
| 1 |
-
---
|
| 2 |
-
license: mit
|
| 3 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
license: mit
|
| 3 |
+
tags:
|
| 4 |
+
- rna
|
| 5 |
+
- biology
|
| 6 |
+
- rna-design
|
| 7 |
+
- biomolecule-design
|
| 8 |
+
- 3d-design
|
| 9 |
+
- inverse-folding
|
| 10 |
+
- inverse-design
|
| 11 |
+
---
|
| 12 |
+
|
| 13 |
+
# gRNAde Datasets
|
| 14 |
+
|
| 15 |
+
[](https://www.biorxiv.org/content/10.1101/2025.11.29.691298)
|
| 16 |
+
[](https://github.com/chaitjo/geometric-rna-design/blob/main/LICENSE)
|
| 17 |
+
[](https://github.com/chaitjo/geometric-rna-design)
|
| 18 |
+
|
| 19 |
+
This repository contains datasets and wet-lab experimental data for **gRNAde**, a generative AI framework for RNA inverse design.
|
| 20 |
+
|
| 21 |
+
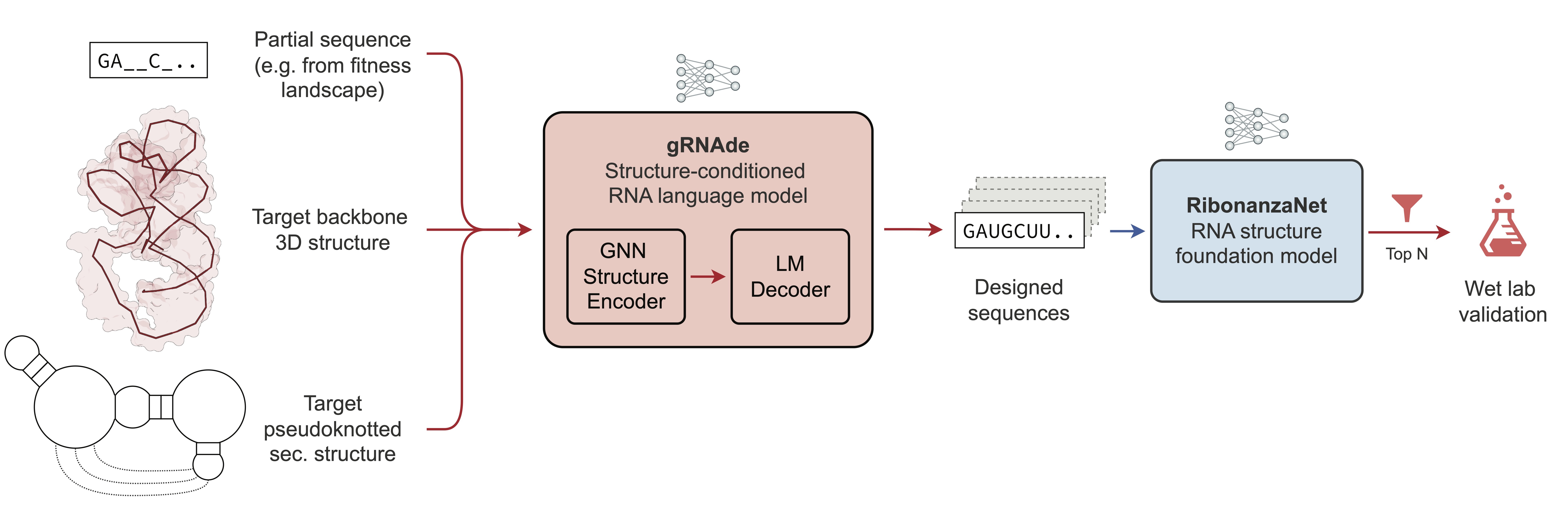
|
| 22 |
+
|
| 23 |
+
## 📦 Datasets and Experimental Data
|
| 24 |
+
|
| 25 |
+
```
|
| 26 |
+
.
|
| 27 |
+
├── RNASolo_31102023_processed.pt # Pre-processed training dataset (ready for ML)
|
| 28 |
+
├── RNASolo_31102023_raw.tar.gz # Raw PDB structures from RNASolo
|
| 29 |
+
└── projects # Data for reproducing design campaigns in the paper
|
| 30 |
+
├── openknot_benchmark # Eterna OpenKnot Benchmark for psuedoknotted RNA design
|
| 31 |
+
└── rna_polymerase_ribozyme # Generative Design of RNA polymerase ribozymes
|
| 32 |
+
```
|
| 33 |
+
|
| 34 |
+
### Pre-processed Training Dataset
|
| 35 |
+
- **File**: `RNASolo_31102023_processed.pt`
|
| 36 |
+
- **Description**: ML-ready dataset of RNA 3D structures from the Protein Data Bank
|
| 37 |
+
- **Source**: RNASolo database (October 31, 2023 cutoff)
|
| 38 |
+
- **Resolution**: ≤4.0 Å
|
| 39 |
+
- **Format**: PyTorch tensors with sequences, 3D coordinates, secondary structures, and metadata
|
| 40 |
+
- **Use case**: Training new gRNAde models or fine-tuning for specific RNA families
|
| 41 |
+
|
| 42 |
+
### Raw PDB Structures
|
| 43 |
+
- **File**: `RNASolo_31102023_raw.tar.gz`
|
| 44 |
+
- **Description**: Raw PDB files for all RNA structures from RNASolo
|
| 45 |
+
- **Contents**: Thousands of experimentally determined RNA structures
|
| 46 |
+
- **Use case**: Custom data processing pipelines or structure visualization
|
| 47 |
+
|
| 48 |
+
### Eterna OpenKnot Competition Data
|
| 49 |
+
- **Directory**: `projects/openknot_benchmark/`
|
| 50 |
+
- **Description**: Designs and experimental validation data from the Eterna OpenKnot Benchmark
|
| 51 |
+
- **Contents**: Chemical probing data, OpenKnot scores, and designed sequences
|
| 52 |
+
- **Targets**: 40 pseudoknotted RNA puzzles including riboswitches, ribozymes, and viral elements
|
| 53 |
+
|
| 54 |
+
### RNA Polymerase Ribozyme Data
|
| 55 |
+
- **Directory**: `projects/rna_polymerase_ribozyme/`
|
| 56 |
+
- **Description**: Functional design campaign data for engineering RNA polymerase ribozymes
|
| 57 |
+
- **Contents**: Generated sequences, activity assays, fitness landscapes
|
| 58 |
+
|
| 59 |
+
## 🚀 Quick Start
|
| 60 |
+
|
| 61 |
+
After setting up the [gRNAde codebase](https://github.com/chaitjo/geometric-rna-design), datasets can be downloaded manually or using HuggingFace CLI (recommended):
|
| 62 |
+
|
| 63 |
+
```bash
|
| 64 |
+
# Ensure you are in the base directory
|
| 65 |
+
cd ~/geometric-rna-design
|
| 66 |
+
|
| 67 |
+
# Install HuggingFace CLI (https://huggingface.co/docs/huggingface_hub/main/en/guides/cli)
|
| 68 |
+
pip install -U "huggingface_hub[cli]"
|
| 69 |
+
# alternate: curl -LsSf https://hf.co/cli/install.sh | bash
|
| 70 |
+
# alternate: brew install huggingface-cli
|
| 71 |
+
|
| 72 |
+
# Download all datasets to data/ directory
|
| 73 |
+
huggingface-cli download chaitjo/gRNAde_datasets --local-dir data/ --repo-type dataset
|
| 74 |
+
```
|
| 75 |
+
|
| 76 |
+
Alternately, download specific files:
|
| 77 |
+
|
| 78 |
+
```bash
|
| 79 |
+
# Download only the pre-processed training dataset
|
| 80 |
+
huggingface-cli download chaitjo/gRNAde_datasets RNASolo_31102023_processed.pt --local-dir data/ --repo-type dataset
|
| 81 |
+
|
| 82 |
+
# Download only raw PDB structures
|
| 83 |
+
huggingface-cli download chaitjo/gRNAde_datasets RNASolo_31102023_raw.tar.gz --local-dir data/ --repo-type dataset
|
| 84 |
+
```
|
| 85 |
+
|
| 86 |
+
## Citations
|
| 87 |
+
|
| 88 |
+
```
|
| 89 |
+
@article{joshi2025generative,
|
| 90 |
+
title={Generative inverse design of RNA structure and function with g{RNA}de},
|
| 91 |
+
author={Joshi, Chaitanya K and Gianni, Edoardo and Kwok, Samantha LY and Mathis, Simon V and Lio, Pietro and Holliger, Philipp},
|
| 92 |
+
journal={bioRxiv},
|
| 93 |
+
year={2025},
|
| 94 |
+
publisher={Cold Spring Harbor Laboratory}
|
| 95 |
+
}
|
| 96 |
+
|
| 97 |
+
@inproceedings{joshi2025grnade,
|
| 98 |
+
title={g{RNA}de: Geometric Deep Learning for 3D RNA inverse design},
|
| 99 |
+
author={Joshi, Chaitanya K and Jamasb, Arian R and Vi{\~n}as, Ramon and Harris, Charles and Mathis, Simon V and Morehead, Alex and Anand, Rishabh and Li{\`o}, Pietro},
|
| 100 |
+
booktitle={International Conference on Learning Representations (ICLR)},
|
| 101 |
+
year={2025},
|
| 102 |
+
}
|
| 103 |
+
|
| 104 |
+
@incollection{joshi2024grnade,
|
| 105 |
+
title={g{RNA}de: A Geometric Deep Learning pipeline for 3D RNA inverse design},
|
| 106 |
+
author={Joshi, Chaitanya K and Li{\`o}, Pietro},
|
| 107 |
+
booktitle={RNA Design: Methods and Protocols},
|
| 108 |
+
pages={121--135},
|
| 109 |
+
year={2024},
|
| 110 |
+
publisher={Springer}
|
| 111 |
+
}
|
| 112 |
+
```
|